
AG7: closing genomes with PacBio |
PacBio CCS reads are long and accurate (error rate around 0.1 %). In addition PacBio provides a very uniform coverage (even for AT rich or GC rich regions) and a random distribution in the pattern of errors along the sequence. Our system AG7 is able to join illumina contigs using PacBio CCS reads.
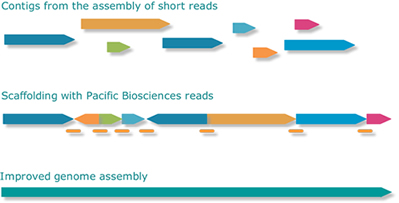
We lengthen illumina contigs and join them based on CCS PacBio reads connecting the ends of two different contigs. The analysis of these connections allows the scaffolding of the illumina contigs and then the final joining of the contigs getting the closing of bacterial genomes.
The strategy of our method is:
- First to assemble the illumina reads, obtaining an assembly that
usually has a high local precision, and then
- Join the illumina contigs using the CCS PacBio long sequences.
The length of these sequences allows us to solve the main problems of the Illumina read assembly that are the repeats, the non-uniform coverage and the non-random error distribution.
The number of PacBio CCS reads needed to get a sufficient coverage for a bacterial genome is very low due to the length of the reads and to the uniform coverage. So it allows the use of innovative methods and algorithms to manage them.
Ask about our prices! Contact us: [email protected]
Digital favorites
- Non Gamstop Casinos
- Migliori Casino Non Aams
- Gioco Aviator
- Non Gamstop Casino
- Non Gamstop Casino
- Non Gamstop Casino
- Casino Italiani Non Aams
- Meilleur Casino En Ligne
- Best Online Casinos
- Non Gamstop Casino Sites UK
- Non Gamstop Casino UK
- Best Non Gamstop Casinos
- Gambling Sites Not On Gamstop
- UK Casinos Not On Gamstop
- Non Gamstop Casino Sites UK
- Casino Not On Gamstop
- Non Gamstop Casinos UK
- Casino Online Non Aams
- UK Casino Not On Gamstop
- UK Casinos Not On Gamstop
- Non Gamstop Casino UK
- Best Casinos Online
- Casino Online Non Aams
- Siti Di Slot Online
- Bitcoin Casino
- Casino En Ligne Crypto
- Site Paris Sportif Belgique
- Migliori App Casino
- Casino En Ligne France
- Meilleur Site De Poker En Ligne
- Paris Sportifs Crypto
- Meilleur Site De Paris Sportif International
- オンラインカジノ アプリ おすすめ
- Meilleur Casino En Ligne France
- Casino En Ligne
- Meilleur Casino En Ligne Français
- オンラインカジノサイト
- Best Crypto Casino
- Casino En Ligne Français
- Casino En Ligne Le Plus Payant
 © Era7 Information Technologies SLU 2012 |
© Era7 Information Technologies SLU 2012 |